
- #Installation of package car had nonzero exit status how to
- #Installation of package car had nonzero exit status install
- #Installation of package car had nonzero exit status iso
GetOption("repos")' replaces Bioconductor standard repositories, see '?repositories' for details replacement repositories: CRAN: Bioconductor version 3.15 (BiocManager 1.30.17), R 4.2.0 ( ucrt) Installing package(s) '38.knownGene' installing the source package ‘38.knownGene’ trying URL ' ' Content type 'application/x-gzip' length 44812729 bytes (42.7 MB) downloaded 42.7 MB The downloaded source packages are in ‘C:\Users\AppData\Local\Temp\RtmpsPAYMJ\downloaded_packages’ Warning message: In install.packages(.) : installation of package ‘38.knownGene’ had non-zero exit status
#Installation of package car had nonzero exit status install
Any ideas how I can install the package? Thanks!īiocManager::install("38.knownGene") Unfortunately, the error message is not so helpful.
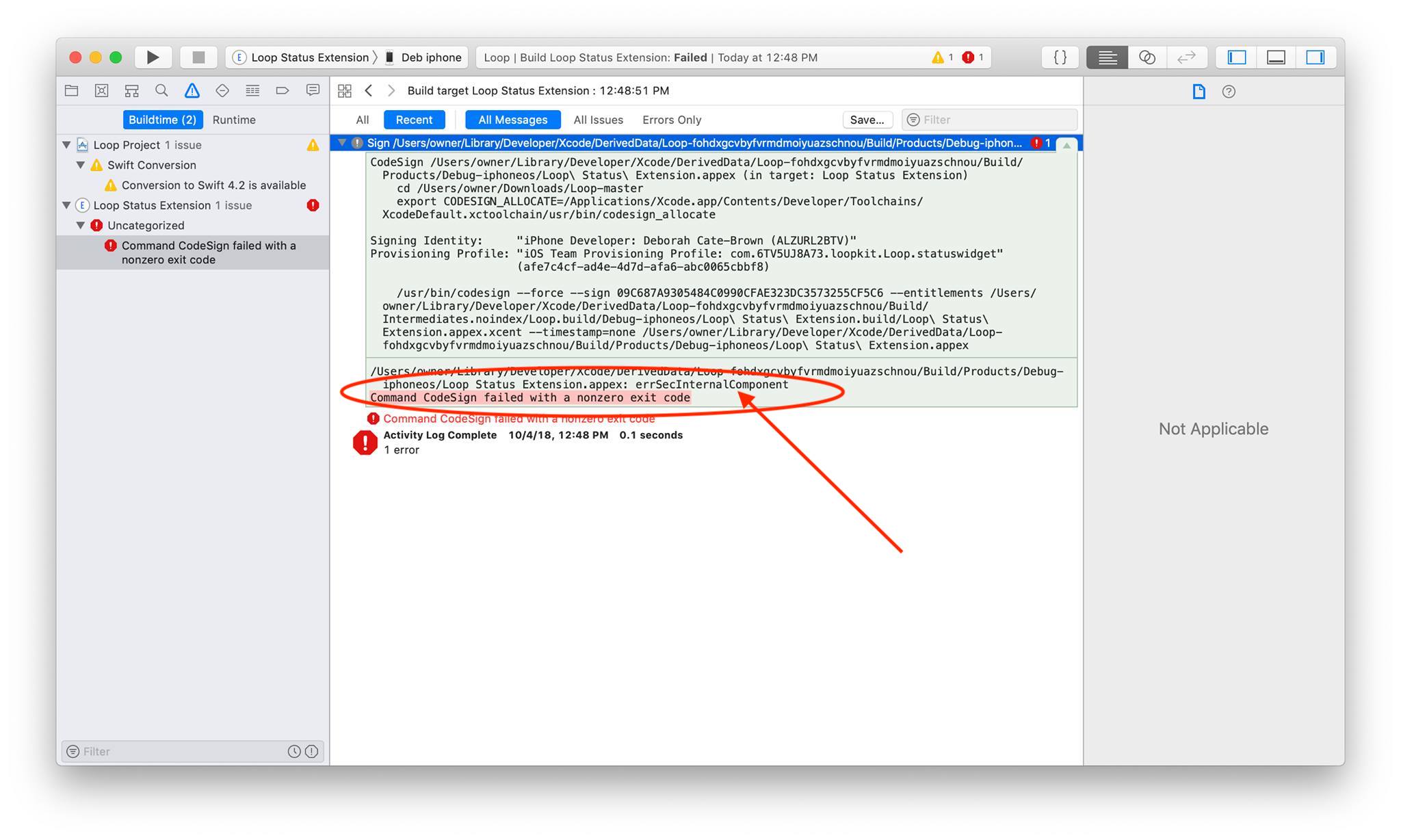
Package fftw3 was not found in the pkg-config search path.
#Installation of package car had nonzero exit status iso
(cached) yesĬhecking for gcc -std=gnu11 option to accept ISO C89. (cached) yesĬhecking whether gcc -std=gnu11 accepts -g. (cached) gcc -std=gnu11Ĭhecking whether we are using the GNU C compiler.
#Installation of package car had nonzero exit status how to
none neededĬhecking how to run the C preprocessor. yesĬhecking for gcc -std=gnu11 option to accept ISO C89.

yesĬhecking whether gcc -std=gnu11 accepts -g. noĬhecking whether we are using the GNU C compiler. a.outĬhecking whether we are cross compiling. yesĬhecking for C compiler default output file name. ** package ‘qqconf’ successfully unpacked and MD5 sums checkedĬhecking whether the C compiler works. Installing package into ‘/PHShome/se22/R/x86_64-pc-linux-gnu-library/4.1’Īlso installing the dependency ‘qqconf’ trying URL ' 'Ĭontent type 'application/x-gzip' length 519510 bytes (507 KB)Ĭontent type 'application/x-gzip' length 1097503 bytes (1.0 MB) Thank you so much in advance for any help! Installation of package ‘mutoss’ had non-zero exit statusĥ: In install.packages("Seurat", dependencies = TRUE) :Īlthough I use R 4.1.2, I am having the same problem. Installation of package ‘hdf5r’ had non-zero exit statusģ: In install.packages("Seurat", dependencies = TRUE) :Ĥ: In install.packages("Seurat", dependencies = TRUE) : Today I've tried a fresh R install and using install('Seurat', dependencies = TRUE), I still got similar errors:ġ: In install.packages("Seurat", dependencies = TRUE) :Ģ: In install.packages("Seurat", dependencies = TRUE) : Then, I've tried install.packages('sn') and got:ĮRROR: dependency ‘mnormt’ is not available for package ‘sn’ Installation of package ‘metap’ had non-zero exit status Installation of package ‘TFisher’ had non-zero exit status Installation of package ‘sn’ had non-zero exit status I followed the instructions, but I got problems with install.packages('metap'): This can be accomplished with the following commands: - install.packages('BiocManager') BiocManager::install('multtest') install.packages('metap'). Its output is the message:Įrror: Please install the metap package to use FindConservedMarkers. However, the function FindConservedMarkers stopped working now (in the same tutorial, I didn't modify anything). I was following the guided tutorial Stimulated vs Control PBMCs using RStudio.


 0 kommentar(er)
0 kommentar(er)
